
Educator Training
We offer up-to-date teacher training through biology workshops and professional development for teachers in genetics and biotechnology. With federal and private foundation funding, we offer these free workshops to middle school, high school, and college educators, especially those in the areas of genetics, biology, genomics, and bioinformatics.
Summer of Nanopore Sequencing 2024
- May 15–17: Spelman College, Atlanta, GA
- May 29–31: Arecibo C3 STEM Center, Arecibo, PR
- June 10–12: Skyline College, San Bruno, CA
- June 17–19: DNA Learning Center NYC at City Tech, Brooklyn, NY
- June 18–20: University of Hawaii at Manoa, Honolulu, HI
- June 20–22: Granite Technical Institute and Natural History Museum of Utah, Salt Lake City, UT
- June 24–26: James Madison University, Harrisonburg, VA
Nanopore sequencing, which analyzes individual DNA molecules in real time, is the “next big thing” in biology education. It promises to put real-time DNA sequencing within reach of the any motivated bioscience teacher. We envision a MinION miniature nanopore sequencing device in every biology teaching lab within a decade.
The DNA Learning Center (DNALC) and Oxford Nanopore are working together to adapt nanopore sequencing for use in education – including improved chemistry, workflows, directions, and packaging/pricing attractive to high school and college faculty. The DNALC’s popular DNA Subway is currently being redeveloped for full mobile use, including a new line for nanopore sequence analysis. Workshop participants will be the first to test this streamlined approach for combining MinION sequence data with the DNA Subway pipeline–providing DNA sequencing and analysis any time, any place, by anyone. The workshop will appeal especially to high school and college faculty who mentor student research or participate in large, distributed projects, such as DNA barcoding (DNALC), SEA-PHAGES (Howard Hughes Medical Institute), and Tiny Earth (University of Wisconsin). At less than $10 per barcode or metabarcode sample and $30 per phage or organelle genome, nanopore sequencing is a speedy and cost-effective alternative to commercial sequencing. Nanopore takes DNA sequencing out of the “black box,” exposing students to every step of the workflow. Workshop participants will have the unique opportunity to immediately sequence and analyze DNA from samples they bring to the workshop. Barcode, metabarcode, and small genome sequencing will be covered. All workshop participants will receive a $300 stipend. Although we expect that most participants will commute to the workshop, funding for travel, room & board is available for a limited number of qualified applicants living outside commuting distance.Supported by grants from the National Science Foundation: Improving Undergraduate STEM Education (#1821657, #2216349), Advanced Technological Education (#1901984), and Arecibo Center for STEM Education and Research (#2321729); and the National Institutes of Health Science Education Partnership Award (#5R25GM137355). Equipment and supplies provided by Oxford Nanopore.
- In-person free workshop
- $300 stipend
- May 15–17: Spelman College, Atlanta, GA
- May 29–31: Arecibo C3 STEM Center, Arecibo, PR
- June 10–12: Skyline College, San Bruno, CA
- June 17–19: DNA Learning Center NYC at City Tech, Brooklyn, NY
- June 18–20: University of Hawaii at Manoa, Honolulu, HI
- June 20–22: Granite Technical Institute & Natural History Museum of Utah, Salt Lake City, UT
- June 24–26: James Madison University, Harrisonburg, VA
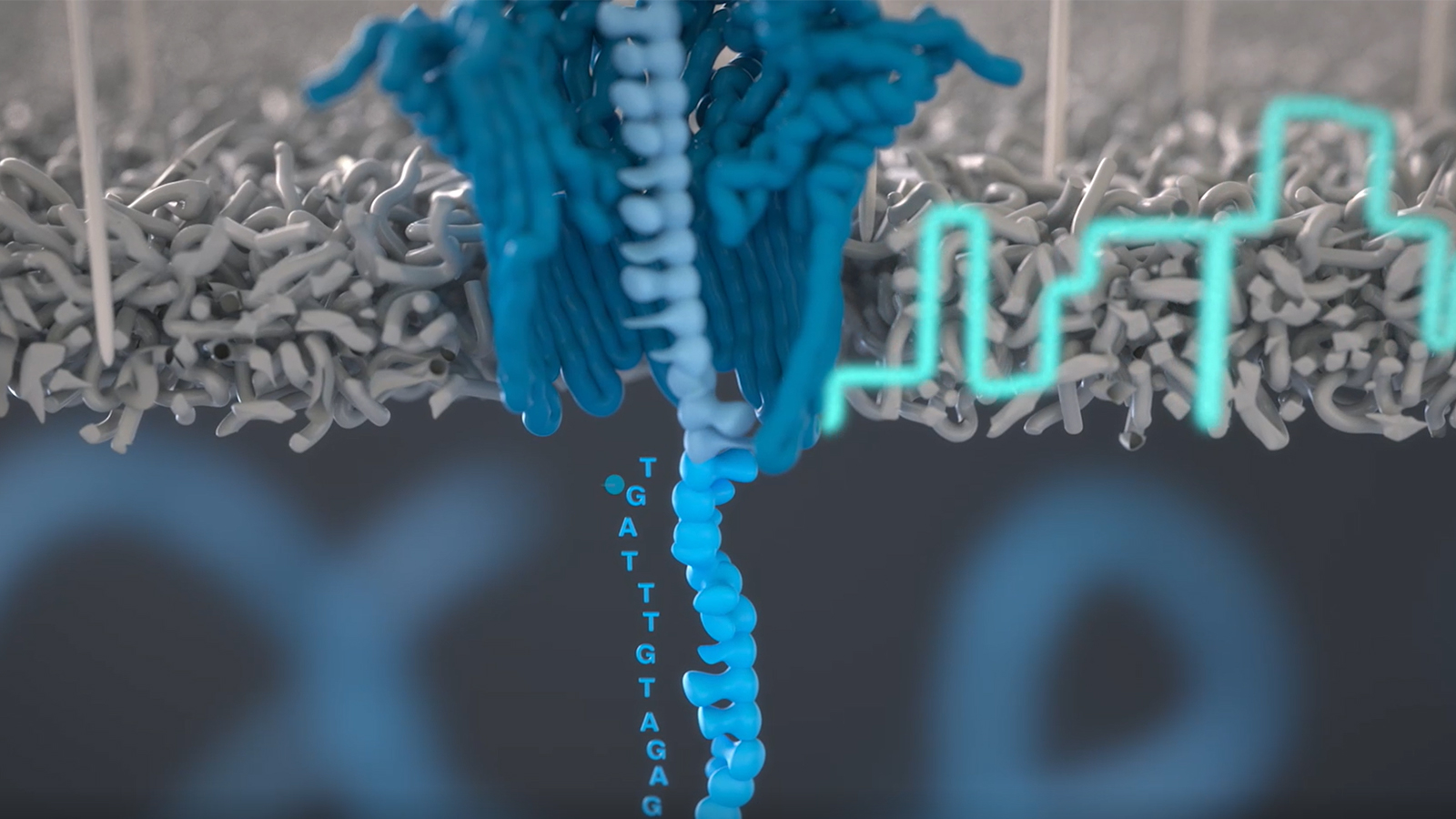

Using Cell-free Systems to Synthesize and Express Bacteriophage Genomes: Practical Biomanufacturing for Bioscience Educators
June 3–6, 2024
DNA Learning Center NYC at City Tech
62 Tillary Street, Brooklyn, New York 11201

Synthetic biology and biomanufacturing are buzzwords of an emerging bioeconomy based on our increasing ability to manipulate living systems. This workshop will equip bioscience educators with the knowledge, skills, and resources to introduce students to a simple workflow to manufacture bacteriophages (phages) with new host specificity, illustrating how biomolecules are adapted to interact with cell-surface receptors.
Join Vincent Noireaux, an instructor of the annual Synthetic Biology course at Cold Spring Harbor Laboratory, to come up-to-the minute with a new, cell-free transcription-translation (TXTL) system he developed to biomanufacture phages in vitro. This workshop emerges from a distinguished lineage of bacteriophage research that began at Cold Spring Harbor Laboratory (CSHL) and provided the first tools to explore the molecular mechanics of living cells. The “Phage Course,” founded at CSHL in 1945 by Max Delbrück and Salvador Luria, trained the first two generations of molecular biologists. Al Hershey and Martha Chase’s “blender experiment,” conducted at CSHL in 1952, provided conclusive evidence that DNA is the molecule of heredity. Delbrück, Luria, Hershey shared the 1969 Nobel Prize for this seminal work.
Bacteriophages are being rediscovered as powerful tools to meet the pharmaceutical challenge of untreatable, multi-drug resistant bacterial infections. In the food industry, bacteriophages prevent formation of biofilms on equipment surfaces, sanitize fresh fruits and vegetables, and extend the shelf life of packaged foods. Using phages to treat bacterial infections in livestock increases resilience and helps eliminate the reservoir of antibiotic-resistant bacteria. Phages also provide a virtually limitless source of bioactive materials that are increasingly exploited in biotechnology, nanotechnology, and bioremediation.
Phages are used in education as examples of simple genetic systems. The SEA-PHAGES Program of the Howard Hughes Medical Institute annotates phage genomes, and is one of the most widely implemented infrastructures for course-based undergraduate research experiences (CUREs). TXTL is a logical next step for students who have been exposed to phage and/or bacterial genetics, providing them an opportunity to explore the use of phages in biomanufacturing.
Workshop participants will conduct hands-on experiments to express reporter genes and whole phage genomes in vitro, using a cell-free extract. Participants will complete an entire workflow to engineer wild-type T7 phage to infect a new E. coli host. This begins with long PCRs to amplify several fragments of the T7 genome, plus PCR mutagenesis of the tail fiber gene. The PCR products are then assembled and packaged as complete T7 genomes using the cell-free TXTL system. Spotting assays compare the host range of wild-type and mutant T7 phages. Participants will then conduct nanopore DNA sequencing, delivering same-day results to investigate point mutations in the tail fiber gene that account for infectivity.
A $500 stipend will be provided. Travel funds are available.
Funded by grants from the National Science Foundation: Future Manufacturing #2228971; Improving Undergraduate STEM Education #1821657
- In-person free workshop
- $500 stipend
- June 3–6, 2024
- DNA Learning Center NYC at City Tech
62 Tillary Street, Brooklyn, New York 11201 - Directions
- Workshop schedule (PDF)
DNA Barcoding Workshop–Citizen DNA Barcode Network
July 29–August 2, 2023, 9:30 a.m.–3:30 p.m. EDT
Dolan DNA Learning Center, Cold Spring Harbor, NY
The Cold Spring Harbor Laboratory DNA Learning Center (DNALC) is offering a five-day, in-person workshop on DNA barcoding for the community-science driven Citizen DNA Barcode Network (CDBN) program. This free workshop is intended for science and nature enthusiasts who are interested in becoming collaborators of CDBN, and CDBN staff at collaborating institutions. High school teachers* on Long Island with an interest in implementing authentic research experiences with students through Barcode Long Island (BLI) are also welcome to apply. Trained mentors interested in a program refresher are also encouraged to attend.
*The Cold Spring Harbor Laboratory DNA Learning Center is an approved Sponsor of Continuing Teacher and Leader Education (CTLE).
Participants in this workshop will learn:
- Introduction to DNA barcoding and biodiversity
- Sample collection and documentation
- DNA extraction
- Polymerase Chain Reaction (PCR)
- Gel electrophoresis
- Bioinformatics–DNA Subway’s Blue Line
- Phylogenetics
- Program implementation
The DNALC developed a Citizen DNA Barcode Network in collaboration with the New York Hall of Science. Just as the unique pattern of bars in a universal product code (UPC) identifies each consumer product, a “DNA barcode” is a unique pattern of DNA sequence that identifies each living thing. Science centers, nature preserves, and other informal science education facilities introduce DNA barcoding and biodiversity to citizen scientists as they obtain DNA sequences to identify potentially harmful, beneficial, and indicator insect species. Program participants provide important species barcode sequence and location information to global biodiversity databases that aim to advance range maps and show how species are responding to environmental change.
Travel and housing will NOT be covered by the DNALC.
- In-person free workshop
- July 29–August 2, 2023
- 9:30 a.m.–3:30 p.m. EDT
- Register by July 19, 2024
- Dolan DNA Learning Center
334 Main St, Cold Spring Harbor, NY 11724 - Directions


Sites of Major DNALC Faculty Workshops, 1985-2014
This map shows the locations of the DNALC's faculty workshops taught over more than thirty years.
Open the map key map key to show/hide the years in groups of three. Click the check boxes to show or hide the years. Click the dots for information on host institution, year, and instructional level of participating faculty. Map can be opened full screen in a separate browser window by clicking the full screen icon at the upper-right.



