Problem 41: DNA is only the beginning for understanding the human genome.
Experiment with gene knock outs.
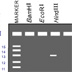
HI! When Susumu Tonegawa and his lab made a strain of mice carrying a floxed NMDA receptor gene, they used a targeting vector that would not disrupt the gene in other cells. This is a simplified version of the NMDA gene; its exons are represented by the red boxes. The gene actually has 22 exons. Which of the following vectors can be used to knock out the NMDA gene when Cre is present? The exons between the two loxP sites are removed by Cre, disabling the NMDA receptor. When Cre is absent, the loxP sites have no effect on the receptor because they sit in the gene's introns. Tonegawa and his lab electroporated this vector into mouse ES cells and then added neomycin. What type of cells were isolated? Only ES cells with a homologous recombination of the vector. (Yes, but this is not the only answer.) Only ES cells with a random recombination of the vector. (Yes, but this is not the only answer.) ES cells with a recombination (homologous or random) of the vector. (That is correct) ES cells without a recombinant vector. (No, ES cells without the vector will die; they are not resistant to neomycin.) Neomycin killed cells that did not integrate the vector, but this did not distinguish between ES cells that homologously or randomly integrated the vector. To pick out the ES cells harboring the homologous recombination, DNA from part of each ES colony was digested with a restriction enzyme. In the wild-type NMDA gene, this generates a 17-kb fragment. In the homologous recombinant, the enzyme cuts within the loxP site and generates a 15-kb fragment. In the homologous recombinant, the enzyme cuts within the loxP site and generates a 15-kb fragment. The fragments were run out on a gel, and a radioactive probe that bound to the third exon was added. Click in the lane on the gel where the vector has homologously recombined into the ES cell. ES cells with the homologously recombined vector will show two restriction pieces. These ES cells, which came from true-bred black mice, were injected into a blastocyst from true-bred white-coated mice. The blastocyst grew into a chimeric mouse. What kinds of sex cells can this chimera produce? The chimera produces three types of sex cells. The cells derived from the blastocyst can only produce sex cells carrying the a allele. The cells derived from the ES cells can produce sex cells carrying the B allele, and sex cells carrying the B allele and the floxed gene. After breeding the chimera to get mice heterozygous for the floxed gene, the heterozygotes were bred with another strain that was heterozygous for floxed as well as for cre. What fraction of the progeny from this cross will be homozygous for the floxed gene and heterozygous for the cre gene? (Hint: use a Punnett square) A Punnett square reveals that 2/16, or 1/8, of the offspring will be homozygous for the floxed gene and heterozygous for cre. Though I/8 of the progeny were the conditional null mutants, the researchers had to use PCR to identify them. Which set of primers can identify the cre gene? Primers are represented by the yellow boxes. PCR amplifies the DNA between the two primers. Because one of the primers binds to the cre gene itself, PCR would not amplify any DNA if the cre gene was missing. CONGRATULATIONS!!! YOU'RE SO SMART!
susumu tonegawa, kb fragment, recombination dna, radioactive probe, restriction enzyme, neomycin, receptor gene, knock outs, exon, introns, vector, fragments, cells, nmda receptor
- ID: 16869
- Source: DNALC.DNAFTB
Related Content
16856. Animation 41: DNA is only the beginning for understanding the human genome.
Mario Capecchi describes proteomics; the large-scale study of protein structure and function. Brian Sauer explains gene knock outs.
16550. Problem 24: The RNA message is sometimes edited.
Map DNA molecules using restriction enzymes.
16725. Animation 35: DNA responds to signals from outside the cell.
James Darnell explains how chemical signals turn eukaryotic genes on and off.
16686. Problem 32: Some DNA can jump.
DNAFTB Problem 32: Explore other organisms with transposable elements.
897. Gene knockout in mice
This method uses homologous recombination to disable a gene of interest to produce a genetic knockout.
16529. Animation 24: The RNA message is sometimes edited.
Rich Roberts and Phil Sharp explain restriction enzymes, electrophoresis, and split genes.
16734. Problem 35: DNA responds to signals from outside the cell.
Explore signal transduction.
16705. Animation 34: Genes can be moved between species.
Stanley Cohen and Herbert Boyer transform bacteria with a recombinant plasmid, and Doug Hanahan studies induced transformation.
15920. DNA arrays
In the 1990s, DNA arrays provided the means to analyze patterns of gene expression at different timepoints in a living cell.
15476. Mechanism of Recombination, 3D animation with with basic narration
Genetic engineering: inserting new DNA into a plasmid vector.